Research
Design, development and testing new multi-target oriented compounds for a treatment and prevention of chronic disorders and diseases (chronic complications in diabetic patients, neurodegenerative diseases, oxidation stress related tissue damage).
Molecular targets: aldo-keto reductases, Ca-ATPase from sarcoplasmic reticulum, cholinesterases, monoamine oxidases, reactive products of oxidation stress.
Chemical and enzyme mechanisms of molecular oxygen activation and tissue damage under hypoxic conditions. Toxic effects of reactive oxygen species and glucose under conditions of hyperglycemia with main attention to diabetic complications; enzyme mechanisms of poyol pathway in the ethiology of diabetic complications. Oxidative injury of Ca-ATPase from sarcoplasmic reticulum as a calcium regulatory enzyme . In silico approaches to antioxidant activity, ligand-protein interaction, QSAR and virtual screening.
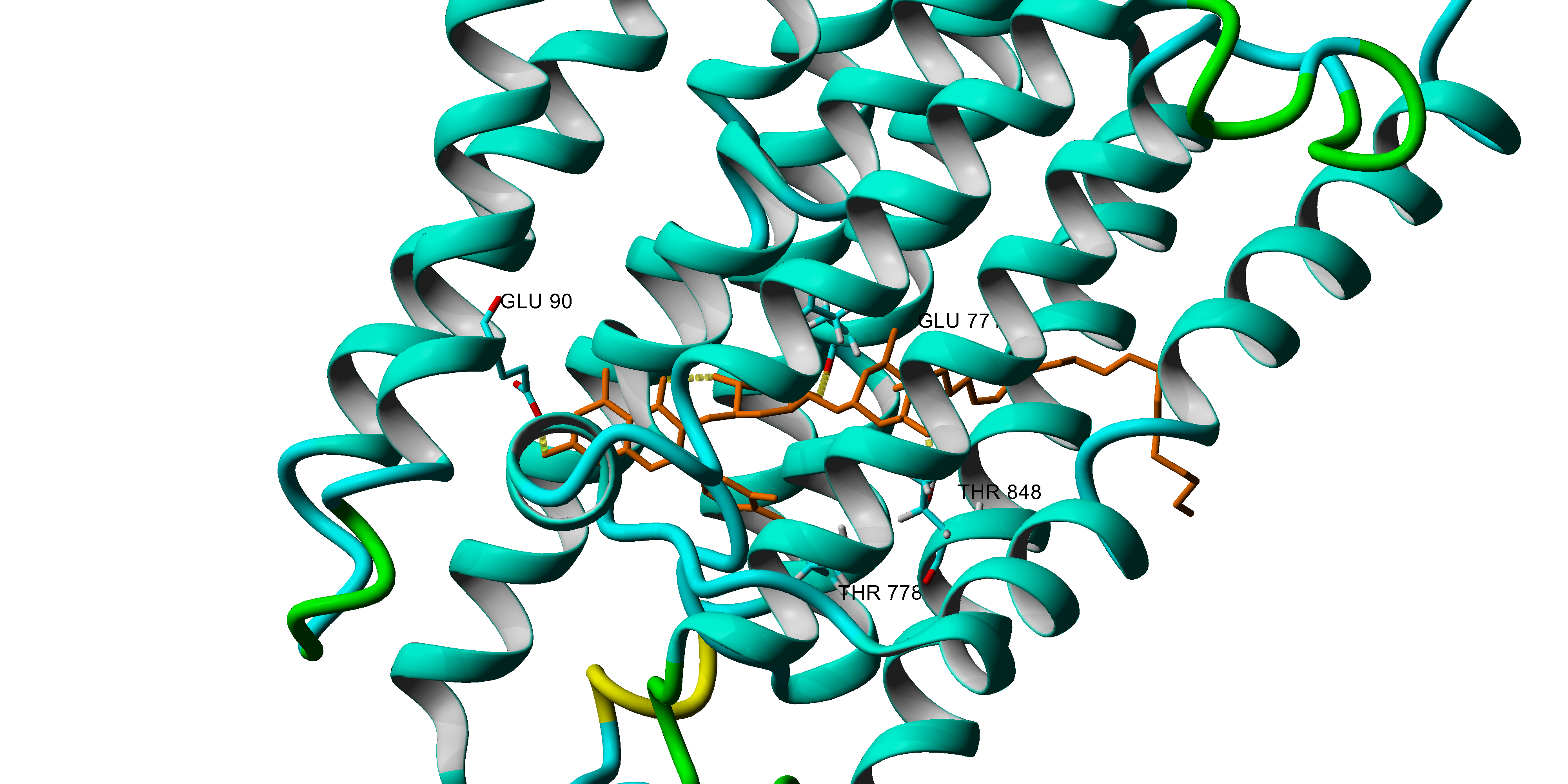
Methods
Chemistry and biochemistry of biotransformation pathways of xenobiotics; inhibition, activation, induction and spectral properties of monooxygenases and reductases. Chemical and physico-chemical structural changes of lipids and proteins initiated by free radical attack. Cell culture techniques in studying cellular damage by oxidative and osmotic stress. Animal models of experimental diabetes and models of hypoxia-ischemia. Quantum chemical and molecular mechanics methods, QSAR, molecular dynamics.
Software for in silico study
Spartan’08 – conformational search (systematic, MC)
QCH calculations
multiprocessing (DFT B3LYP 6-31G*)
YASARA – protein modeling, docking,
AMBER, YASARA, YAMBER3, NOVA FF
molecular dynamics, homology modeling
multiprocessing
DRAGON - calculation of molecular descriptors > 3000
prediction of therapeutic properties
STATISTICA – statistical analysis
GAUSSIAN 09 – exact QCH calculations, HPC klaster STU
MOE – molecular modeling, combinatorial libraries, MD,
pharmacophore constructing, chemical and molecular mechanics methods,
QSAR, molecular dynamics
NAMD - molecular dynamics, AUREL in Computing Centre SAS
